MMB1116 Quick and Clean Cloning
MMB1116: Quick and Clean Cloning
2014/01/09 22:18:27
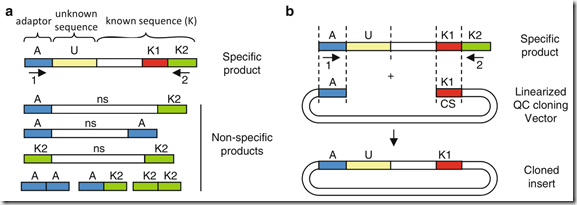
Fig. 1 Principle of the QC cloning strategy.
( a ) DNA fragments containing known and unknown flanking sequences are amplified by PCR using a primer homologous to an adaptor sequence (primer 1 and sequence A) attached to the end of the unknown sequence (U) and a primer homologous to a region of the known sequence (primer 2 and region K2). In addition to specific products, PCR amplification can yield nonspecific products (ns) and primer dimers.
( b ) Fragments are cloned via homology between sequences present in both the insert and the vector: sequences A and sequence K1 (also called the catching sequence, CS). Since primer 2 used for PCR amplification of the insert does not overlap with region K1, nonspecific products and primer dimers cannot be cloned
Abstract
Quick and Clean Cloning
doi: 10.1007/978-1-62703-764-8_3
Identification of unknown sequences that flank known sequences of interest requires PCR amplification of DNA fragments that contain the junction between the known and unknown flanking sequences. Since amplified products often contain a mixture of specific and nonspecific products, the quick and clean (QC) cloning procedure was developed to clone specific products only. QC cloning is a ligation-independent cloning procedure that relies on the exonuclease activity of T4 DNA polymerase to generate single-stranded extensions at the ends of the vector and insert. A specific feature of QC cloning is the use of vectors that contain a sequence called catching sequence that allows cloning specific products only. QC cloning is performed by a one-pot incubation of insert and vector in the presence of T4 DNA polymerase at room temperature for 10 min followed by direct transformation of the incubation mix in chemo-competent Escherichia coli cells.