How to Select Candidate Genes for your Association Study?
How to Select Candidate Genes for your Association Study?
2011/09/07 22:54:10
A candidate gene is a gene that is suspected of being involved in a diseases or specific trait. When we talk about candidate genes association studies, we actually mean that we want to analyze some mutations in the candidate genes that we have selected for our study. So first, how can we choose candidate genes for a genetic study? There are many different ways to do so.
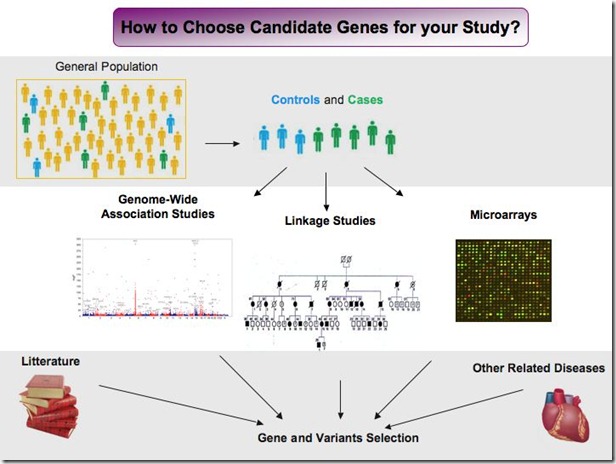
- One of the most common is to select a gene which codes for a protein that has been implicated in the disease, or which codes for other proteins in the same pathway. For example, if you are studying hypertension, you could choose to focus on the angiotensin-converting enzyme (ACE) that has been linked to hypertension in many studies in human and animal models, and you can also choose to study proteins involved in the renin-angiotensin-aldosterone system (such as the AGT), which is a pathway where the ACE plays an important role. Similarly, you could also test genes that have been associated with related diseases. Following the same example, you could check the genes implicated in stroke or myocardial infarction.
- Another widely used approach is to use the data provided by linkage studies. Linkage studies scan thousand of markers in the entire genome and look for peak of association with a disease. Typically these studies have been performed in large families affected by a disease. Once a peak region is found, a deeper screening of the DNA region can be performed. Sometimes it can be difficult to screen the entire region, so people usually select some candidate genes in the region identified and choose to analyze mutations in those genes. So a way to start a new study is to review all linkage studies available in your disease and check carefully which genes have been identified in the linkage regions. You can then check the literature for more information on these genes and choose a few of them for your own project.
- Microarrays are another popular way to select candidate genes. Microarrays are gene expression studies where the expression of genes is compared between cases and controls. The approach permits to identify hundreds of genes that have an altered expression in patients with the disease, which can be upregulated or downregulated. The problem is that it is impossible to know if the expression is altered as a result of the disease or if it is actually causing the disease. In any case, microarrays are great to do a pre-selection of candidate genes to analyze in your samples. You can thus browse microarray studies or perform one yourself in a few samples, and use it as a base for deeper genetic analysis. One option for example is to focus on all genes that show at least 5x more expression in your patients compared to your controls.
- Finally, one of the newest approaches to identify candidate genes are GWAS or genome-wide analysis studies. These studies screen the entire genome for millions of genetic markers and can reveal the association of hundreds of different mutations with your disease. It is then very difficult to know which ones are really true or important for the pathology of the disease, but it is a good way to select some mutations and genes and to study them in your population. A deep sequencing of the regions where a lot of mutations are identified could for example reveal new genes or new mutations.
Of course, all these approaches can also be used in combination. Here is a slide I made to try to sum up the different ways to find candidate genes: