【PNAS】新技术实现纳米孔内快速测序DNA
PNAS:新技术实现纳米孔内快速测序DNA
2010/08/25 07:49:19
有望实现1000美元测序个人基因组
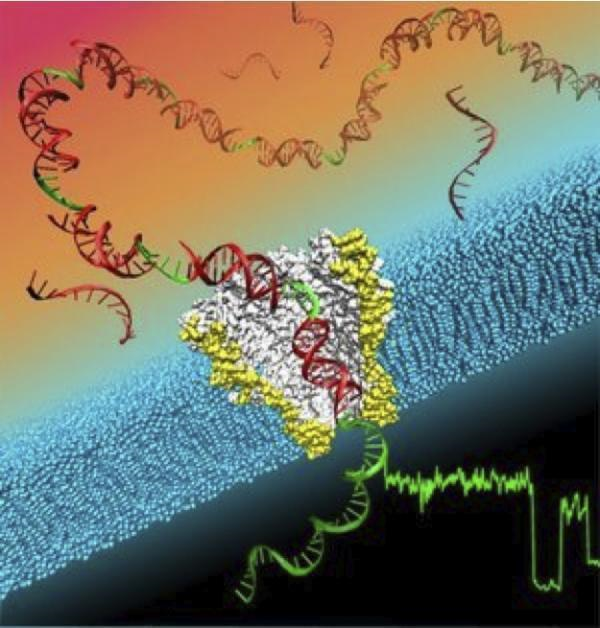
This illustration depicts a single strand of DNA moving through a nanopore that is being used to sequence the DNA. (Credit: Image courtesy of University of Washington)
据《每日科学》近日报道,由美国华盛顿大学物理学家领导的研究小组设计了一种新技术,可在纳米孔内对DNA进行快速测序,而且价格比较便宜。新方法可为癌症、糖尿病或某些成瘾患者量身绘制个性化基因测序蓝图,提供更加高效的个体医疗。相关论文发表于美国《国家科学院院刊》(PNAS)。
论文主要作者、华盛顿大学物理教授简斯·冈德拉克表示,他们结合了生物和纳米技术,研制出这种DNA阅读器,阅读器内纳米微孔使用了一种取自耻垢分支杆菌的细胞外膜孔道蛋白A。这种纳米微孔只有1个纳米大小,仅够用来测量一个DNA的单分子链。
研究人员把微孔放在一层浸泡在氯化钾溶液中的膜上,并施加一个小的电压,让电流通过微孔。不同的核苷酸通过纳米微孔时,回路中的电流就会随之改变,这些电流称为特征信号。胞核嘧啶、鸟嘌呤、腺嘌呤和胸腺嘧啶这些DNA的基本组成要素,会生成不同的特征信号。
研究小组解决了两个主要问题,一是生成仅容一条DNA单链通过的纳米微孔,且每次只能通过一个DNA分子。伯明翰亚拉巴马州立大学的迈克尔·涅德维斯改良了细菌,生成了合适的微孔。第二个问题是让核苷酸以每秒100万个的速率通过纳米微孔,冈德拉克说,这实在太快了,阅读器还无法在这种速度下对每个DNA分子信号分类整理。为了解决这一点,研究人员在每个要测量的核苷酸之间附带了一段双链DNA,双链DNA在微孔中流动不那么顺畅,磕磕绊绊地通过微孔,便可将下一个通过微孔的单链延迟几毫秒。这种延迟尽管只有千分之几秒,电信号却有了充足时间来识别目标核苷酸,从而从示波器轨迹上准确读出这些DNA序列。
这项研究由美国国家卫生研究院和美国人类基因研究院资助,旨在降低成本,使人类基因组完整测序成本降到1000美元或更少。该研究始于2004年,当时完整测序一个人的基因要花费1000万美元,而新的测序技术使人们向1000美元测序的目标迈进了一大步。
Nanoscale DNA Sequencing Could Spur Revolution in Personal Health Care
ScienceDaily (Aug. 21, 2010) — In experiments with potentially broad health care implications, a research team led by a University of Washington physicist has devised a method that works at a very small scale to sequence DNA quickly and relatively inexpensively.
That could open the door for more effective individualized medicine, for example providing blueprints of genetic predispositions for specific conditions and diseases such as cancer, diabetes or addiction.
“The hope is that in 10 years people will have all their DNA sequenced, and this will lead to personalized, predictive medicine,” said Jens Gundlach, a UW physics professor and lead author of a paper describing the new technique published the week of Aug. 16 in the Proceedings of the National Academy of Sciences.
The technique creates a DNA reader that combines biology and nanotechnology using a nanopore taken from Mycobacterium smegmatis porin A. The nanopore has an opening 1 billionth of a meter in size, just large enough to measure a single strand of DNA as it passes through.
The scientists placed the pore in a membrane surrounded by potassium-chloride solution. A small voltage was applied to create an ion current flowing through the nanopore, and the current’s electrical signature changed depending on the nucleotides traveling through the nanopore. Each of the nucleotides that are the essence of DNA – cytosine, guanine, adenine and thymine – produced a distinctive signature.
The team had to solve two major problems. One was to create a short and narrow opening just large enough to allow a single strand of DNA to pass through the nanopore and for only a single DNA molecule to be in the opening at any time. Michael Niederweis at the University of Alabama at Birmingham modified the M. smegmatis bacterium to produce a suitable pore.
The second problem, Gundlach said, was that the nucleotides flowed through the nanopore at a rate of one every millionth of a second, far too fast to sort out the signal from each DNA molecule. To compensate, the researchers attached a section of double-stranded DNA between each nucleotide they wanted to measure. The second strand would briefly catch on the edge of the nanopore, halting the flow of DNA long enough for the single nucleotide to be held within the nanopore DNA reader. After a few milliseconds, the double-stranded section would separate and the DNA flow continued until another double strand was encountered, allowing the next nucleotide to be read.
The delay, though measured in thousandths of a second, is long enough to read the electrical signals from the target nucleotides, Gundlach said.
“We can practically read the DNA sequence from an oscilloscope trace,” he said.
Besides Gundlach and Niederweiss, other authors are Ian Derrington, Tom Butler, Elizabeth Manrao and Marcus Collins of the UW; and Mikhail Pavlenok at Alabama-Birmingham.
The work was funded by the National Institutes of Health and its National Human Genome Research Institute as part of a program to create technology to sequence a human genome for $1,000 or less. That program began in 2004, when it cost on the order of $10 million to sequence a human-sized genome.
The new research is a major step toward achieving DNA sequencing at a cost of $1,000 or less.
“Our experiments outline a novel and fundamentally very simple sequencing technology that we hope can now be expanded into a mechanized process,” Gundlach said.